CRISPR is a gene-editing technology that is used to selectively modify DNA to achieve gene knockout, knock-in, activation, or repression through targeted removal, addition or alteration of disease-implicated sequences [1,2]. The technology is based on a naturally-occurring genome-editing defense system found in bacteria, called Clustered Regularly Interspaced Short Palindromic Repeats, from which it gets its name [1,2]. As shown in Figure 1, CRISPR leverages an RNA-guide protein that “scans” the genome, looking for a precise DNA sequence that it has been “programmed” to target [1,2]. It then cuts the target DNA with an enzyme (most famously Cas9) and initiates repair using genetic material that has been altered to have a therapeutic effect [1,2].
Editing the Genome
The CRISPR gene-editing system uses an enzyme called Cas9 and a customized guide RNA to help target, cut, alter, and stitch up particular stretches of the genome. It can disrupt a gene or edit in a new one.
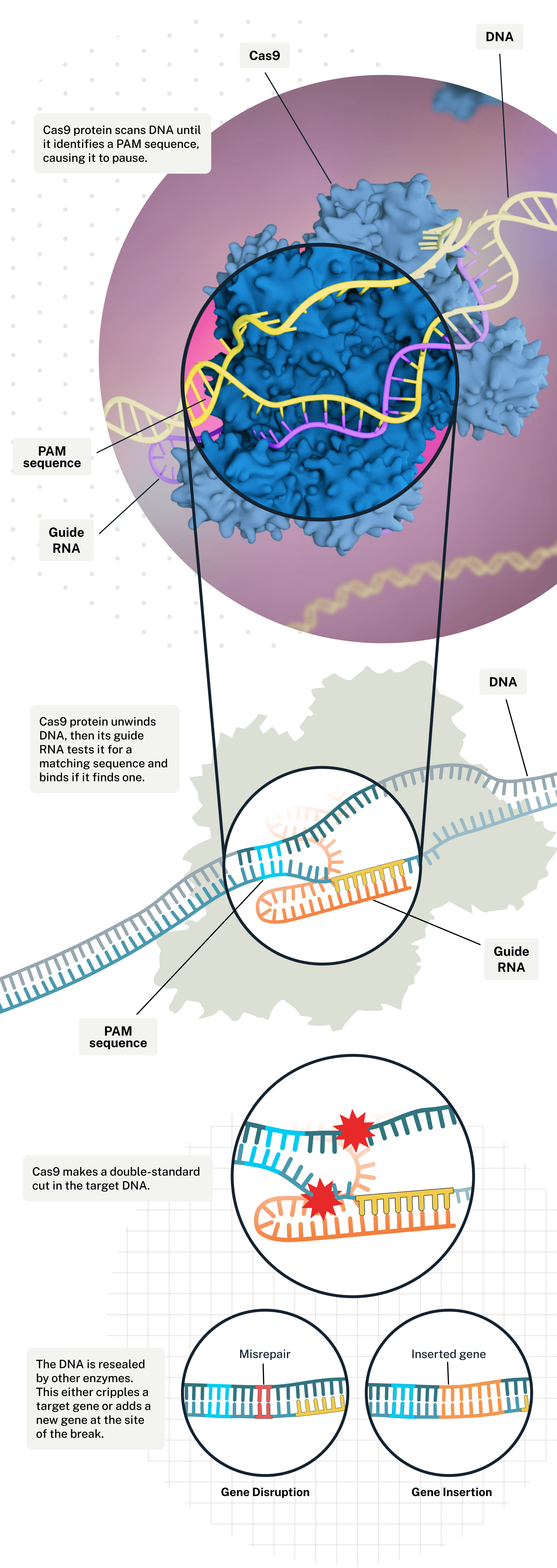
Figure 1: Editing the genome with the CRISPR Cas-9 gene-editing system.
(Image credit: Illustration by Falconieri Visuals. From Scientific American).
Recent CRISPR Advances
After Dr. Jennifer Doudna and Dr. Emmanuelle Charpentier won the 2020 Nobel Prize in Chemistry for their CRISPR-Cas9 “genetic scissors,” expectations for the technology’s potential grew [3]. And, as detailed by the Innovative Genomics Institute’s 2023 clinical trial update, CRISPR is currently being studied for a variety of uses, including [4]:
Direct editing of problematic genes: CRISPR is making great progress in relatively easy-to-target genetic conditions, such as inherited blood disorders and genetic blindness. In fact, Vertex Pharmaceuticals expects a regulatory decision on its CRISPR-based sickle-cell-disease treatment by the end of 2023 [5].
Novel gene editing approaches: Researchers are also exploring novel approaches for CRISPR, from reprogramming existing drugs to target new diseases to developing non-viral delivery systems (including lipid nanoparticles that have an affinity for the liver) to performing edits without double-strand DNA breaks.
Pathogen-targeted treatments: Treatments are being studied that would directly target the genome of hard-to-treat pathogens, such as those implicated in chronic urinary tract infections and HIV.
Cell-therapy modifications: Researchers are using CRISPR to expand the possibilities of cell therapies; for example, in several areas of cancer research, teams are looking to evolve CAR-T treatments beyond individualized patient-derived treatments by using CRISPR to create and edit cell therapies from healthy donors that could be used in wider populations. CRISPR editing is also being explored as a way to reduce immune response in type-1 diabetes patients treated with stem-cell-derived pancreatic cells.
Beyond its application in therapeutic gene editing, CRISPR is also being used as a diagnostic tool; for example, Sherlock Biosciences is developing CRISPR technology that can be used to detect the unique genetic fingerprints encoded in virtually any DNA or RNA sequence in any organism or pathogen [6].
Key Challenges Impeding CRISPR’s Targeted Success
CRISPR has made impressive progress considering that it has only been pursued as a therapeutic for just over a decade. But a more rapid revolution and application beyond simple targets and single-gene disorders is being impeded by two key issues—off-target effects and delivery challenges.
Off-Target Effects of CRISPR Guide RNA
CRISPR primarily depends on guide RNA (gRNA) to precisely pinpoint the locations on a particular gene sequence where modifications should occur. A critical consideration in this process is the precision of the guide RNA and its ability to bind/influence unintended gene locations. When designing therapies directed at addressing genetic disease variants, the potential for off-target effects constantly looms as a significant concern. An important dimension of mitigating this concern involves an up-close look at the sequence data during the design phase. To minimize these risks, meticulous design of gRNAs and repair templates becomes imperative, often facilitated by molecular design tools like SnapGene.
Take, for example, the CRISPR-Cas9 system. It uses gRNA that comprises a 20 base-pair target sequence next to a protospacer-adjacent motif (PAM site) to direct the Cas9 endonuclease to the cleavage site. While a number of Cas9 variants have been developed to reduce off-target activity, there is still some risk. The onus is on researchers to thoroughly scan for gRNA CRISPR sites in a desired gene (or selected region) and carefully search for potential off-target binding sites. Each gRNA must be scored according to how many off-target sites it will potentially bind to and how similar the off-target sites are to the original sequence. As shown in Figure 2, sequence analysis software like Geneious Prime can help researchers accomplish this. Because gRNAs are 20 nucleotides long, the potential off-targets are limited to closely related sequences, hence off-site cleavage is relatively predictable and potentially avoidable. However, several factors can impact how accurately the gRNA directs CRISPR effector protein cleavage. So to be more comprehensive in evaluating potential off-target damage, there are several techniques available that can scan the genome for mismatches. They vary in sensitivity and specificity, and no one technique is 100% conclusive. In some cases, teams need to complete whole-genome sequencing for absolute certainty.
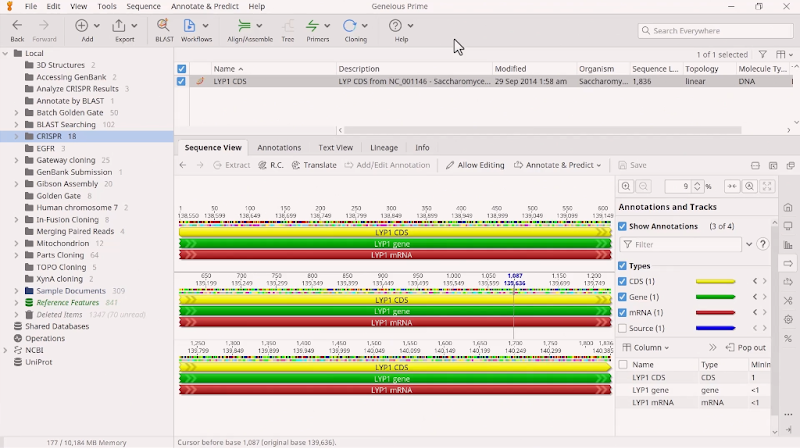
Figure 2: Geneious Prime can be used to find and annotate CRISPR sites in a sequence and check for off-target binding sites.
CRISPR Delivery Challenges
Another significant limitation to CRISPR is the delivery of CRISPR components to an intended target. This limitation is more pronounced in complex eukaryotic systems and in therapeutics, where delivery needs to be optimized to certain cell types while minimizing potential toxic side effects. When trying to develop therapies that have targets within hard-to-access cells or tissues, this problem is especially complex.
Target access is critical because if the CRISPR-Cas9 system can’t bind to the target DNA sequence, it can’t make the needed cleavage. This is especially difficult in eukaryotic cells, where DNA is tightly packaged with proteins and other molecules. There are a number of factors that need to be considered, which affect target access; these include factors such as DNA-binding proteins, sequence-specific effects, and the structure of chromatin.
Chromatin is the complex of DNA, proteins, and other molecules that make up our chromosomes. The structure of chromatin varies depending on the cell type and the gene being expressed. Tightly packed chromatin can make it difficult for CRISPR-Cas9 to access the target DNA sequence, because the intended cut site is buried within the complex. In addition to target access, there are thousands of DNA-binding proteins that bind to DNA, like transcription factors and histones, DNA repair proteins, chromatin remodelers, and many others. These proteins can and do compete with CRISPR-Cas9 for binding to the target DNA sequence. Lastly, there are sequence-specific effects where the sequence of the target DNA sequence itself can affect target access. For example, CRISPR-Cas9 may have difficulty binding to sequences that are rich in GC base pairs.
It’s for these reasons we think most of the CRISPR R&D to date has focused on “easy to access” targets.
Viral delivery systems have been a common means for directing CRISPR components to cells and tissues of interest, but even then, issues arise, like carrying-capacity restrictions, the potential to trigger negative immune responses, and tissue-specificity limitations [7]. Researchers are increasingly exploring non-viral alternatives, like lipid nanoparticle delivery systems or chemical modifications, which may improve delivery without increasing the risk of negative immune responses [4,8-11].
Overcoming CRISPR Design Hurdles
To address these key R&D hurdles, researchers are increasingly working at higher throughput to study multiple gRNA candidates at once, as well as collaborating across disciplines to explore how chemical modification might help. Unfortunately, as detailed below, doing such is hard for teams whose fragmented informatics solutions make sharing data and optimizing cross-discipline workflows a struggle.
Large Batch Guide RNA Design in CRISPR
Large batch gRNA design can help uncover the candidates with the lowest potential for off-target effects, which is particularly necessary when working on complex multifactorial diseases, where more than one gene or genomic region needs to be targeted. Many researchers are now working in a high-throughput fashion, designing and assessing hundreds of guides at a time. And although these teams are casting a wider net, they still need to thoroughly interrogate all potential guides for off-target effects and efficacy. Batch discovery of CRISPR sites on sequence lists or groups of sequence documents creates an abundance of data that must be scored for activity and specificity; researchers may also want to find optimal pairs and build off-target databases to search against, as well as design repair templates, predict the effects of editing on protein translation, and potentially analyze sequencing results from CRISPR editing experiments. Doing this is daunting when relying on standalone speciality applications that aren’t integrated tightly to a united R&D platform that serves as a master repository for all results and insights.
Chemical Modifications in CRISPR
As mentioned, another way teams are attempting to expand the use of the CRISPR-Cas9 system is by exploring the potential of chemical modifications, including those made to the gRNA or Cas9 system. Chemical modification holds potential to improve target-binding affinity and nuclease resistance, as well as reduce off-target effects and immunogenicity [8-11]. Researchers at Case Western Reserve University are even studying whether chemical alterations could help control not only the location of CRISPR modifications, but also the timing [11]. But, as we discussed in our blog “Top 5 Challenges for Collaborative Chemically-Modified Biologics R&D,” cross-discipline collaboration between biology and chemistry teams is difficult when those teams think and work in different ways, with different workflows, tools, and data types. A solution that supports the unique needs of both sides can make a world of difference.
Collaborative Cross-Discipline CRISPR R&D with Dotmatics
R&D teams must be able work together to select and analyze gRNA, study off-target effects, explore the impact of repair-template editing, design delivery vectors, and test various chemical modifications. Dotmatics supports large-scale collaborative CRISPR R&D by uniting an array of functionality within a user-friendly and data-centric R&D platform, including:
CRISPR Guide RNA Design Tools - Use speciality tools, such as Geneious or SnapGene, to design, score, and optimize gRNA to target the specific segments of DNA that need gene editing. Accelerate efforts with batch discovery and scoring of CRISPR sites on sequence lists or groups of sequence documents.
Molecular Editing and Sequence Analysis Software - Design repair templates and perform analysis tasks, such as PCR and DNA sequence analysis, sequence alignment, and gene expression analysis, using leading molecular biology and bioinformatics software Geneious and SnapGene.
Vector Design and Chemical Modification Tools - Design viral vectors that optimize delivery to target sites using Geneious or SnapGene. Use Dotmatics chemistry tools to explore chemical modifications or non-viral delivery systems that may help reach difficult target cells and tissues and reduce immunogenicity.
Inventory and Registration Systems - Manage the cells, clones, and samples used in experiments with a registration system that accommodates biologic entities with or without chemical modifications.
Flow Cytometry Analysis Software - Measure the impact of gene editing on cell populations using flow cytometry software, such as OMIQ or FCS Express.
Electronic Laboratory Notebooks - Track and log all project details with personalizable ELNs that offer flexible support for cross-discipline CRISPR-design workflows.
CRISPR R&D teams choose Dotmatics because it:
Supports cross-discipline collaboration - Unite CRISPR design tools like Geneious and SnapGene on an end-to-end R&D platform, creating a single source of scientific truth, even when biologists and chemists are collaborating on chemical-modification.
Optimizes individual productivity - Reduce constant back-and-forth between software; for example users can directly register gRNAs from the ELN, Geneious or SnapGene, or registration system.
Accelerates innovation - Uncover insights and empower decision-making by easily collating all relevant R&D data and using advanced analysis tools, such as Prism, to analyze, graph and present data exactly as needed.
Accommodates flexibility - Think and work outside the box with tools like Dotmatics ELN, which has been optimized to support the fluidity and flexibility needed to push the boundaries of possibility for CRISPR therapeutics.
Learn More About Dotmatics for CRISPR R&D
Contact us to learn more about the ways Dotmatics can support your CRISPR therapeutic gene editing projects. Or, dig deeper into Dotmatics CRISPR screening and design tools by:
References
What is CRISPR? [Webpage]. Innovative Genomics Institute. Accessed September 20, 2023.
What are genome editing and CRISPR-Cas9? [Webpage]. Medline Plus. Accessed September 20, 2023.
The Nobel Prize in Chemistry 2020. [Press release]. The Royal Swedish Academy of Sciences. October 7, 2020.
Henderson, H. CRISPR Clinical Trials: A 2023 Update. Innovative Genomics Institute. March 17, 2023.
Newman, C. FDA sets decision dates for Vertex, CRISPR gene editing drug. BioPharmaDive. June 9, 2023.
What is Sherlock? [Webpage]. Sherlock Biosciences. Accessed September 20, 2023.
Xu CL, Ruan MZC, Mahajan VB, Tsang SH. Viral Delivery Systems for CRISPR. Viruses. 2019 Jan 4;11(1):28. doi: 10.3390/v11010028.
Qiubing Chen, Ying Zhang, Hao Yin. Recent advances in chemical modifications of guide RNA, mRNA and donor template for CRISPR-mediated genome editing. Advanced Drug Delivery Reviews. 2021 Jan; 168;246-258. https://doi.org/10.1016/j.addr.2020.10.014.
Rozners, E. Chemical Modifications of CRISPR RNAs to Improve Gene-Editing Activity and Specificity. J. Am. Chem. Soc. 2022, 144, 28, 12584–12594. https://pubs.acs.org/doi/10.1021/jacs.2c02633
Zhang H, Kelly K, Lee J, et. al. Self-delivering CRISPR RNAs for AAV Co-delivery and Genome Editing in vivo. bioRxiv [Preprint]. 2023 Mar 23:2023.03.20.533459. doi: 10.1101/2023.03.20.533459.
Gene editing improved with chemical process: Case Western Reserve University researchers combine novel chemical method with CRISPR gene-editing tools to target disease-specific versions of genetic code. The Daily. Case Western Reserve University. March 16, 2022.



